
>>SR Microscopy
Related Products
Nano-Cyte™LC
Nano-View®/M Series
Nano-View Series®
Nano-LPS Series
Nano-LP Series
Nano-BioS Series
Nano-Bio Series
Nano-T Series
Nano-Z Series
Nano-F Series
C-Focus
Nano-F3D
Questions?
 Super-resolution (SR) microscopy is a broad category of recently developed methods in fluorescence-based light microscopy that have cleverly side-stepped the resolution limit imposed by diffraction on lens based optical systems. These super-resolution (SR) imaging methods show great promise as a research tool for basic biological research as they have the theoretical potential to achieve resolutions equaling those of electron microscopy, while also being far less expensive and less invasive.
Super-resolution (SR) microscopy is a broad category of recently developed methods in fluorescence-based light microscopy that have cleverly side-stepped the resolution limit imposed by diffraction on lens based optical systems. These super-resolution (SR) imaging methods show great promise as a research tool for basic biological research as they have the theoretical potential to achieve resolutions equaling those of electron microscopy, while also being far less expensive and less invasive. The Nobel Prize in Chemistry for 2014 will be awarded to Eric Betzig, Stefan W. Hell, and William E. Moerner for the development of super-resolved fluorescence microscopy. SR microscopy was chosen as the “Method of the Year” for 2008 by Nature Methods, based on its tremendous potential for increasing our understanding of cellular biology. SR microscopy methods have already achieved resolutions at the scale of 10s of nanometers, producing images of cellular and macromolecular structures with resolutions approaching those attained by electron microscopy. SR microscopy methods are compatible with whole cells, or even live cells, as well as much more cell-structure-friendly fixation methods. Furthermore, they can be used for multiplex labeling with very high molecular specificity, and are also potentially much more economical and simpler to implement. For these and other reasons, it is widely expected that over the next several years, SR microscopy will lead to very significant advances in our knowledge of cellular processes down to the molecular level.
SR microscopy is based on fluorescence microscopy, which involves using specific fluorescent molecular probes within a sample that are capable of absorbing photons of one wavelength and then subsequently emitting photons of another (usually longer) wavelength. This can be done in either wide-field, or by scanning the sample. Appropriate filters are used to eliminate contamination by unwanted wavelengths of light during imaging. The extension of fluorescent microscopy into the SR realm depends on critical properties intrinsic to some fluorescent molecules: the ability to “switch” or “activate” them from one fluorescent state to another using light of specific wavelengths. These processes are known as photo-switching and photo-activation, respectively; and they can be used to proactively manipulate the absorption and emission properties of the fluorescent probes in the sample. SR microscopy exploits these properties to either spatially restrict, or temporally separate the photons emitted by two closely spaced fluorescent sources so that they can be imaged separately, and in doing so circumvents the diffraction limit. The emerging SR microscopy techniques have spurred an alphabet soup of acronyms: STED, RESLOFT, GSD, SSIM, fPALM, PALM, & STORM, to name only some of them. While these methods differ in their details and implementation, each of them takes advantage of particular properties in fluorescent probes, and they have been collectively termed “super-resolution” to indicate that they achieve resolutions beyond the diffraction limit. Please click on a heading below to learn more about super resolution microscopy techniques and how Mad City Labs products can be used in SR microscopy applications. Click the heading a second time to collapse the section. Image at top left: SR microscopy (STORM) image of Cy5-Cy3 labeled tubulin from A431 cells. Image courtesy of Prof. K. Lidke, University of New Mexico. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||
 |
SR Microscopy Techniques | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| Image at left from York AG, Ghitani A, Vaziri A, Davidson MW, Shroff H. Confined activation and subdiffractive localization enables whole-cell PALM with genetically expressed probes. Nature Methods, Vol. 8, No. 4. (13 April 2011), pp. 327-333. Figure 5(b). | |||||||||||||||||||||||||||||||||||||||||||||||||||||
|
PALM is Photo-activated Localization Microscopy, fPALM is fluorescence Photo-activated Localization Microscopy, and STORM is Stochastic Optical Reconstruction Microscopy. PALM, fPALM, and STORM are “localization-based” techniques that take advantage of the photo-activation and photo-switching properties of fluorescent probes in a manner that differs from STED and SSIM. If two point sources of light are emitting photons of the same wavelength simultaneously, the equation of Ernst Abbe given above still governs the resolving power of any microscope. However, if these point sources can be made to emit their photons individually, at different times so that they can be separately collected, the diffraction limit can be circumvented. In localization-based SR imaging, the fluorophore labels are switched on and off stochastically in sparse subsets while imaging these subsets en masse using an EMCCD camera. This is done in the wide-field, rather than by scanning the lasers. Super-resolution is achieved by localizing each fluorescent emitter by computing the center of the point spread function (PSF) of each one as measured by its photon density count on an array of pixels on the EMCCD camera. The final image is built from a stack of many such images, each representing only a small fraction of the total number of the individual fluorescent emitters in the entire field. Localization-based SR microscopy techniques have already achieved resolutions in the realm of 10-20nm in the x-y plane and may have the potential to achieve even higher resolutions. STED is Stimulated Emission Depletion Microscopy. STED microscopy is one of the methods that spatially restricts fluorophore excitation and emission. In this technique, the excitation volume is reduced through the use of two lasers. The first laser excites the fluorophores, while the second laser turns the fluorophores off. By surrounding the first laser spot by a second one with a donut-shaped intensity profile, and adjusting the intensities these two lasers appropriately, the excitation volume can be reduced below the diffraction limit. This technique is fundamentally a scanning-based method, and the excitation and depletion lasers are moved rapidly through the sample while the emitted photons are collected, frame-by-frame, by an electron-multiplying charge-coupled device (EMCCD) camera as this process proceeds. These frames are then assembled into a complete image of the entire field. GSD (ground state depletion) and RESOLFT (reversibly saturable optical fluorescence transitions) are fundamentally similar to STED. STED has produced images of biological samples with lateral resolutions in the 60nm range. SSIM is Saturated Structured Illumination Microscopy. SSIM is a technique that illuminates the sample with wide-field patterned light, and the excitation pattern interacts with the sample’s spatial information resulting in Moiré fringe patterns being produced. The information contained within these fringes can be computationally extracted, and an image of the sample can be calculated using this information that goes beyond the diffraction limit. SSIM has attained resolutions in the 50-100nm range. | |||||||||||||||||||||||||||||||||||||||||||||||||||||
 |
New Developments in SR Microscopy at Mad City Labs | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| Image at left of Mad City Labs Nano-LPS Series XYZ nanopositioning stage | |||||||||||||||||||||||||||||||||||||||||||||||||||||
Stay up to date with new developments by following Mad City Labs' Super-Resolution Microscopy blog. .jpg) Nano-Cyte® is a 3D image stabilization system for microscopy. With Nano-Cyte® you no longer need to be concerned with temperature gradients, sample drift, and microscope drift. Unprecedented stability in the nanometer regime allows the extension of single molecule techniques into the realm of cell biology. Nano-Cyte® is a 3D image stabilization system for microscopy. With Nano-Cyte® you no longer need to be concerned with temperature gradients, sample drift, and microscope drift. Unprecedented stability in the nanometer regime allows the extension of single molecule techniques into the realm of cell biology.
Join the Nano-Cyte® Revolution! | |||||||||||||||||||||||||||||||||||||||||||||||||||||
 |
Recommended Systems for SR Microscopy | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| Image at left of Mad City Labs Nano-F Series objective nanopositioner | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||
 |
Examples of Mad City Labs Systems in SR Microscopy Research | ||||||||||||||||||||||||||||||||||||||||||||||||||||
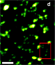
| Image at left from Q. Li, S. S. H. Wu, and K. C. Chou. Sub-diffraction-limit Two-photon Fluorescence Microscopy for GFP-tagged Cell Imaging. Biophys. J . 97, 3224 (2009). Figure 2(d) | |||||||||||||||||||||||||||||||||||||||||||||||||||||
Below are selected publications from customers: Carnegie Mellon University, Chemistry and Molecular Biosensors and Imaging Center, Laboratory for Fluorescence Innovation in Biology
École Polytechnique Fédérale de Lausanne (EPFL), Laboratory of Nanoscale Biology (LBEN)
Harvard University, Chemistry, Zhuang Lab
National Institutes of Health, National Institute of Biomedical Imaging and Bioengineering, Section on High Resolution Optical Imaging
Yale University, Department of Biological & Biomedical Sciences, Bewersdorf Lab
| |||||||||||||||||||||||||||||||||||||||||||||||||||||
Copyright © 2024